SWAN/SIOC: Alignment Between the SWAN and SIOC Ontologies
W3C Interest Group Note 20 October 2009
- This version:
- http://www.w3.org/TR/2009/NOTE-hcls-swansioc-20091020/
- Latest version:
- http://www.w3.org/TR/hcls-swansioc/
- Editor:
- Alexandre Passant, DERI Galway at the National University of Ireland, Galway, Ireland <alexandre.passant@deri.org>
- Paolo Ciccarese, Massachusetts General Hospital / Harvard Medical School <paolo.ciccarese@gmail.com>
- Contributors:
- John G. Breslin, DERI Galway at the National University of Ireland, Galway, Ireland <john.breslin@deri.org>
- Tim Clark, Massachusetts General Hospital / Harvard Medical School <tim_clark@harvard.edu>
Copyright © 2008 W3C® (MIT, ERCIM, Keio), All Rights Reserved. W3C liability, trademark and document use rules apply.
Abstract
This notes describes the alignment between the SWAN - Semantic Web Applications in Neuromedicine - and SIOC - Semantically-Interlinked Online Communities - ontologies, providing a complete model to represent Scientific Discourse in online communities at different levels of granularity (discourse elements and content items).
The goal of this alignment is to make the discourse structure and component relationships much more accessible to computation, so that information can be navigated, compared and understood in context far better that at present, across and within domains.
Status of This Document
This section describes the status of this document at the time of its publication. Other documents may supersede this document. A list of current W3C publications and the latest revision of this technical report can be found in the W3C technical reports index at http://www.w3.org/TR/.
This W3C Interest Group Note describes how one can use the Semantic Web to express and integrate scientific data. These techniques can be used for modeling any data, and the benefits of integration and model consistency apply to other diverse, distributed data domains. It is hoped that this document will inspire further contributions to the ongoing work at Neurocommons and the Health Care and Life Sciences Interest Group, as well as inspire those in other domains to exploit the Semantic Web.
Background for this document is provided by two companion documents:
Semantic Web Applications in Neuromedicine (SWAN) Ontology describes the SWAN ontology for expressing scientific discourse, and
SIOC, SIOC Types and Health Care and Life Sciences describes the SIOC ontology for semantically-interlinked online communities.
This document describes the the use of SWAN and SIOC together to model discourse within scientific communities.
The document was produced by the Semantic Web in Health Care and Life Sciences Interest Group (HCLS), part of the W3C Semantic Web Activity (see charter). Comments may be sent to the publicly archived public-semweb-lifesci@w3.org mailing list.
Publication as an Interest Group Note does not imply endorsement by the W3C Membership. This is a draft document and may be updated, replaced or obsoleted by other documents at any time. It is inappropriate to cite this document as other than work in progress.
The disclosure obligations of the Participants of this group are described in the charter.
1. Introduction
The SWAN (Semantic Web Applications in Neuromedicine) [SWAN-ONT-JBI] project attempts to model Scientific Discourse about Alzheimer's disease and its supporting evidence in a rich and extensible way that is compatible with the way the domain of Alzheimer's research functions as a technology-mediated knowledge ecosystem.
In addition, as described in [SIOC-HCLS], the SIOC (Semantically-Interlinked Online Communities) ontology [SIOC-Specification] can be efficiently used in the Health Care and Life Sciences (HCLS) context as a means to represent the various interactions happening in these communities and their related content in a machine-readable way thanks to Semantic Web technologies.
While not focused on the same objectives, SWAN and SIOC act in a complementary way: SWAN provides fine-grained modeling of scientific discourse elements while SIOC can represent more generic contributions in online communities.
In this note, we present the alignment between the SWAN (version 1.2 [SWAN-HCLS]) and SIOC ontologies, that provides a complete model (based on Semantic Web technologies) to represent Scientific Discourse in online communities at different levels of granularity (discourse elements and content items), a field particularly relevant to the HCLS domain.
2. The SWAN/SIOC Module
2.1. Overview of the SWAN/SIOC mappings
In order to define the alignments between the SWAN and SIOC ontologies, a special module for SIOC has been created (the SWAN/SIOC Module).
This module contains the different statements defining the alignments (subclasses, subproperties, etc.) between the original ontologies in a machine-readable form, i.e. RDF.
This SWAN/SIOC Module is available at http://rdfs.org/sioc/swan.
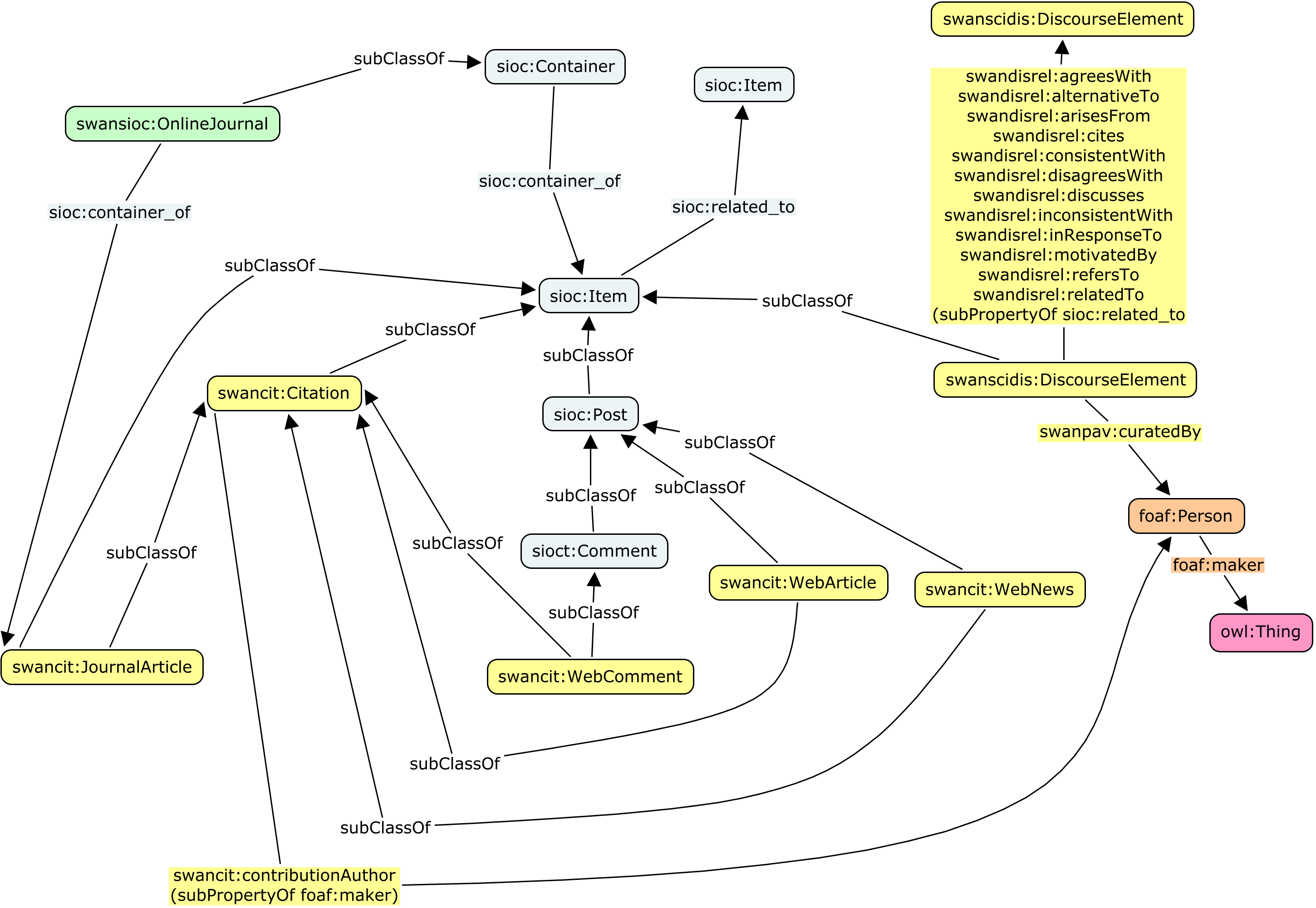
The following figure identifies the various relationships defined in this module, which are also detailed in the remainder of this note.
2.1. Alignment between classes
In order to provide mappings between the classes defining discourse elements (from SWAN, in particular those in the SWAN Citations Ontology and the
SWAN Scientific Discourse Module) and more generic Social Web contributions (from SIOC), the following mappings have been defined.
The subclass relationships are defined using the rdfs:subClassOf property.
Since sioc:Post and sioct:Comment are subclasses of sioc:Item, the six classes from SWAN mentioned in the previous table are all subclasses of sioc:Item.
In addition, a class called swansioc:OnlineJournal has been created in the SWAN/SIOC Module for representing journal-type websites where immutable articles are published and comments are allowed on these articles. This class can be used to represent online publications venues such as StemBook.
This class has been defined as a subclass of sioc:Container, so that a swanscit:JournalArticle can be linked to its container (swansioc:OnlineJournal) using the sioc:has_container property (inverse sioc:container_of).
2.1. Alignment between properties
In addition to the previous classes, mappings have been defined between different properties of the SWAN Scientific Discourse Relationships Ontology and the SIOC Core Ontology.
The following table identifies the defined relationships.
The subproperty relationships are defined using the rdfs:subPropertyOf property.
As one can see when observing these mappings, some of them are redundant. Since they are subproperties of swandisrel:relatedTo, all the previous properties would also become subproperties of sioc:related_to by inference. However, we define such mappings because (1) SIOC applications, that could benefit from these mappings, do not necessarily implement full reasoning and transitive closure over rdfs:subClassOf, and (2) the required computation time when querying data could also benefit from these direct mappings depending on the implementation.
3. Use case
Let us consider once again the ACME Pharma use case that we described in [SIOC-HCLS]. In addition to exposing SIOC information from different services within a company (blogs, wikis, microblogging) in order to integrate them, ACME tools provide a means to let users, when replying to other posts or creating new content (such as wiki pages), to indicate the relationships with other data on the network. Fine-grained modeling of these relationships is provided thanks to SWAN, in particular by using the various properties defined by the SWAN Discourse Relationships Ontology [SWAN-HCLS].
Thanks to the mappings presented in the previous section, as soon as two posts are linked together by such a relationship, a sioc:related_to link is inferred between both, hence enabling distributed conversations on the company network. From a blog post, a user is able to identify that there are several related wiki pages and blog posts. In addition, it is not just these links that can be viewed as sioc:related_to links (in order, for example, to provide a high-level view of a conversation at first glance), but the original SWAN links can also be used to enable more fine-grained navigation, providing ways to zoom in to conversations based on specific relationships.
As an example, when looking at posts about a particular drug treatment, Alice, a researcher at ACME, can immediately identify all related posts (using sioc:related_to links). Then, she may decide to select only the ones disagreeing with the post (via the swandisrel:disagreesWith property) to check if the treatment is appropriate or not. In addition, by using the SIOC Types Module and other properties of SIOC items, she can decide to filter the items so as to browse only the blog posts created by Bob, since she trusts him as an eminent researcher in the domain.
4. Conclusion
In this note, we described the alignment between the SWAN and SIOC ontologies, published via SIOC's SWAN/SIOC Module.
Future direction should include applications of the SWAN/SIOC Module, notably through the Science Collaboration Framework as well as dedicated applications enabling search and navigation from several SWAN knowledge bases.
In addition and following the same idea of unifying discourse representation models to provide better integration of existing data, other mappings between existing RDFS/OWL models for argumentative discussion and scientific discours may be considered, based on models such as aTags [ATAGS], CiTO [CITO], IBIS [IBIS], SALT [SALT] or [SchoolOnto].
For instance, aTags use a subset of the SIOC vocabulary to represent single statements and efforts are under way to represent hundreds of thousands of biomedical statements formalized this way [ATAGS-DATA].
A1. Namespaces used in this document
A2. References
- [ATAGS]
-
Matthias Samwald.
aTags: Associative Tags.
http://hcls.deri.org/atag/
- [ATAGS-DATA]
-
List of datasets in aTag format. See http://esw.w3.org/topic/HCLSIG_BioRDF_Subgroup/aTags/datasets
- [CITO]
-
David Shotton.
CiTO, the Citation Typing Ontology, and its use for annotation of reference lists and visualization of citation networks.
In Proceedings of Bio-Ontologies 2009: Knowledge in Biology.
Ontology available at http://purl.org/net/cito/
- [IBIS]
-
Danny Ayers.
IBIS Vocabulary: Issue-Based Information Systems for the Semantic Web.
http://hyperdata.org/xmlns/ibis/
- [SALT]
-
Tudor Groza, Siegfried Handschuh, Knud Möller and Stefan Decker.
SALT - Semantically Annotated LaTeX for Scientific Publications.
Proceedings of the 4th European Semantic Web Conference (ESWC 2007).
http://salt.semanticauthoring.org/
- [SchoolOnto]
-
The SchoolOnto project.
http://projects.kmi.open.ac.uk/scholonto/
- [SIOC-HCLS]
-
Alexandre Passant (editor).
SIOC, SIOC Types and Health Care and Life Sciences.
W3C HCLS IG Editor's Draft.
http://www.w3.org/2001/sw/hcls/notes/sioc
- [SIOC-Specification]
-
Uldis Bojars, John G. Breslin (eds.).
SIOC Core Ontology Specification.
Evolving document.
http://rdfs.org/sioc/spec
- [SWAN-HCLS]
-
Paolo Ciccarese (editor).
Semantic Web Applications in Neuromedicine (SWAN) Ontology.
http://www.w3.org/2001/sw/hcls/notes/swan/
- [SWAN-ONT-JBI]
-
Paolo Ciccarese, Elizabeth Wu, June Kinoshita, Gwen Wong, Marco Ocana, Alan Ruttenberg, Tim Clark.
The SWAN Biomedical Discourse Ontology.
J Biomed Inform. 2008 Oct;41(5):739-51. Epub 2008 May 4.
A3. Acknowledgements
We would like to particularly thank (in alphabetical order) John Breslin, Scott Marshall and Susie Stephens for their valuable comments and feedback during the writing and editing of this note. We also thank Eric Prud'hommeaux for the subversion magic and technical assistance with the creation of this document, Matthias Samwald for his input regarding aTags, David Shotton for his input regarding CiTO as well as the entire Scientific Discourse Task Force, and more generally the Health Care and Life Science Interest Group within the W3C.